Gaussian Processes Barycentres
In this notebook we'll give an implementation of . In this work, the existence of a Wasserstein barycentre between a collection of Gaussian processes is proven. When faced with trying to average a set of probability distributions, the Wasserstein barycentre is an attractive choice as it enables uncertainty amongst the individual distributions to be incorporated into the averaged distribution. When compared to a naive mean of means and mean of variances approach to computing the average probability distributions, it can be seen that Wasserstein barycentres offer significantly more favourable uncertainty estimation.
import typing as tp
import jax
# Enable Float64 for more stable matrix inversions.
from jax import config
import jax.numpy as jnp
import jax.random as jr
import jax.scipy.linalg as jsl
from jaxtyping import install_import_hook
import matplotlib.pyplot as plt
import numpyro.distributions as npd
from examples.utils import use_mpl_style
config.update("jax_enable_x64", True)
with install_import_hook("gpjax", "beartype.beartype"):
import gpjax as gpx
from gpjax.parameters import Parameter
key = jr.key(123)
# set the default style for plotting
use_mpl_style()
cols = plt.rcParams["axes.prop_cycle"].by_key()["color"]
Background
Wasserstein distance
The 2-Wasserstein distance metric between two probability measures and quantifies the minimal cost required to transport the unit mass from to , or vice-versa. Typically, computing this metric requires solving a linear program. However, when and both belong to the family of multivariate Gaussian distributions, the solution is analytically given by
where and .
Wasserstein barycentre
For a collection of measures , the Wasserstein barycentre is the measure that minimises the average Wasserstein distance to all other measures in the set. More formally, the Wasserstein barycentre is the Fréchet mean on a Wasserstein space that we can write as
where is a weight vector that sums to 1.
As with the Wasserstein distance, identifying the Wasserstein barycentre is often an computationally demanding optimisation problem. However, when all the measures admit a multivariate Gaussian density, the barycentre has analytical solutions
Identifying is achieved through a fixed-point iterative update.
Barycentre of Gaussian processes
It was shown in that the barycentre of a collection of Gaussian processes such that can be found using the same solutions as in . For a full theoretical understanding, we recommend reading the original paper. However, the central argument to this result is that one can first show that the barycentre GP is non-degenerate for any finite set of GPs i.e., . With this established, one can show that for a -dimensional finite Gaussian distribution , the Wasserstein metric between any two Gaussian distributions converges to the Wasserstein metric between GPs as .
In this notebook, we will demonstrate how this can be achieved in GPJax.
Dataset
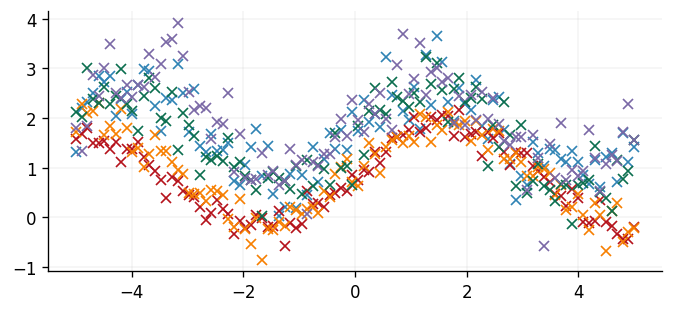
We'll simulate five datasets and develop a Gaussian process posterior before identifying the Gaussian process barycentre at a set of test points. Each dataset will be a sine function with a different vertical shift, periodicity, and quantity of noise.
n = 100
n_test = 200
n_datasets = 5
x = jnp.linspace(-5.0, 5.0, n).reshape(-1, 1)
xtest = jnp.linspace(-5.5, 5.5, n_test).reshape(-1, 1)
f = lambda x, a, b: a + jnp.sin(b * x)
ys = []
for _ in range(n_datasets):
key, subkey = jr.split(key)
vertical_shift = jr.uniform(subkey, minval=0.0, maxval=2.0)
period = jr.uniform(subkey, minval=0.75, maxval=1.25)
noise_amount = jr.uniform(subkey, minval=0.01, maxval=0.5)
noise = jr.normal(subkey, shape=x.shape) * noise_amount
ys.append(f(x, vertical_shift, period) + noise)
y = jnp.hstack(ys)
fig, ax = plt.subplots()
ax.plot(x, y, "x")
plt.show()
Learning a posterior distribution
We'll now independently learn Gaussian process posterior distributions for each dataset. We won't spend any time here discussing how GP hyperparameters are optimised. For advice on achieving this, see the Regression notebook for advice on optimisation and the Kernels notebook for advice on selecting an appropriate kernel.
def fit_gp(x: jax.Array, y: jax.Array) -> npd.MultivariateNormal:
if y.ndim == 1:
y = y.reshape(-1, 1)
D = gpx.Dataset(X=x, y=y)
likelihood = gpx.likelihoods.Gaussian(num_datapoints=n)
posterior = (
gpx.gps.Prior(
mean_function=gpx.mean_functions.Constant(), kernel=gpx.kernels.RBF()
)
* likelihood
)
nmll = lambda p, d: -gpx.objectives.conjugate_mll(p, d)
opt_posterior, _ = gpx.fit_scipy(
model=posterior,
objective=nmll,
train_data=D,
trainable=Parameter,
)
latent_dist = opt_posterior.predict(xtest, train_data=D)
return opt_posterior.likelihood(latent_dist)
posterior_preds = [fit_gp(x, i) for i in ys]
Optimization terminated successfully.
Current function value: -31.740071
Iterations: 10
Function evaluations: 16
Gradient evaluations: 16
Optimization terminated successfully.
Current function value: 30.775673
Iterations: 12
Function evaluations: 19
Gradient evaluations: 19
Optimization terminated successfully.
Current function value: -102.543998
Iterations: 10
Function evaluations: 18
Gradient evaluations: 18
Optimization terminated successfully.
Current function value: -143.031733
Iterations: 12
Function evaluations: 22
Gradient evaluations: 22
Current function value: -271.573651
Iterations: 10
Function evaluations: 57
Gradient evaluations: 45
/home/runner/work/GPJax/GPJax/.venv/lib/python3.11/site-packages/scipy/optimize/_minimize.py:779: OptimizeWarning: Desired error not necessarily achieved due to precision loss.
res = _minimize_bfgs(fun, x0, args, jac, callback, **options)
Computing the barycentre
In GPJax, the predictive distribution of a GP is given by a
TensorFlow Probability
distribution, making it
straightforward to extract the mean vector and covariance matrix of each GP for
learning a barycentre. We implement the fixed point scheme given in (3) in the
following cell by utilising Jax's vmap operator to speed up large matrix operations
using broadcasting in tensordot.
def sqrtm(A: jax.Array):
return jnp.real(jsl.sqrtm(A))
def wasserstein_barycentres(
distributions: tp.List[npd.MultivariateNormal], weights: jax.Array
):
covariances = [d.covariance_matrix for d in distributions]
cov_stack = jnp.stack(covariances)
stack_sqrt = jax.vmap(sqrtm)(cov_stack)
def step(covariance_candidate: jax.Array, idx: None):
inner_term = jax.vmap(sqrtm)(
jnp.matmul(jnp.matmul(stack_sqrt, covariance_candidate), stack_sqrt)
)
fixed_point = jnp.tensordot(weights, inner_term, axes=1)
return fixed_point, fixed_point
return step
With a function defined for learning a barycentre, we'll now compute it using the
lax.scan operator that drastically speeds up for loops in Jax (see the
Jax documentation).
The iterative update will be executed 100 times, with convergence measured by the
difference between the previous and current iteration that we can confirm by
inspecting the sequence array in the following cell.
weights = jnp.ones((n_datasets,)) / n_datasets
means = jnp.stack([d.mean for d in posterior_preds])
barycentre_mean = jnp.tensordot(weights, means, axes=1)
step_fn = jax.jit(wasserstein_barycentres(posterior_preds, weights))
initial_covariance = jnp.eye(n_test)
barycentre_covariance, sequence = jax.lax.scan(
step_fn, initial_covariance, jnp.arange(50)
)
L = jnp.linalg.cholesky(barycentre_covariance)
barycentre_process = npd.MultivariateNormal(barycentre_mean, scale_tril=L)
Plotting the result
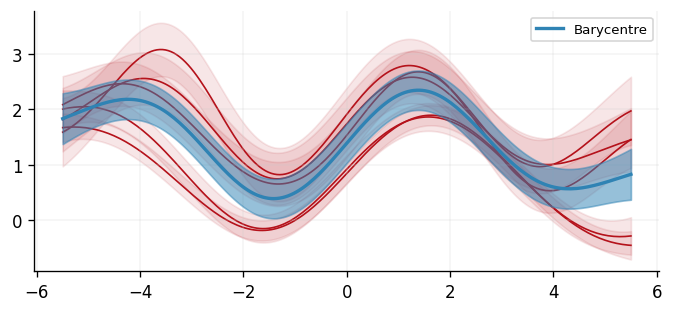
With a barycentre learned, we can visualise the result. We can see that the result looks reasonable as it follows the sinusoidal curve of all the inferred GPs, and the uncertainty bands are sensible.
def plot(
dist: npd.MultivariateNormal,
ax,
color: str,
label: str = None,
ci_alpha: float = 0.2,
linewidth: float = 1.0,
zorder: int = 0,
):
mu = dist.mean
sigma = jnp.sqrt(dist.variance)
ax.plot(xtest, mu, linewidth=linewidth, color=color, label=label, zorder=zorder)
ax.fill_between(
xtest.squeeze(),
mu - sigma,
mu + sigma,
alpha=ci_alpha,
color=color,
zorder=zorder,
)
fig, ax = plt.subplots()
[plot(d, ax, color=cols[1], ci_alpha=0.1) for d in posterior_preds]
plot(
barycentre_process,
ax,
color=cols[0],
label="Barycentre",
ci_alpha=0.5,
linewidth=2,
zorder=1,
)
ax.legend()
<matplotlib.legend.Legend at 0x7f5f69109450>
Displacement interpolation
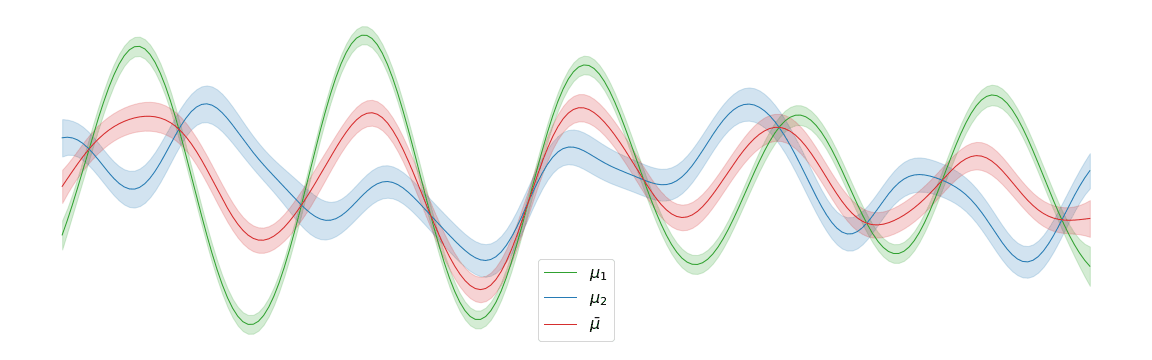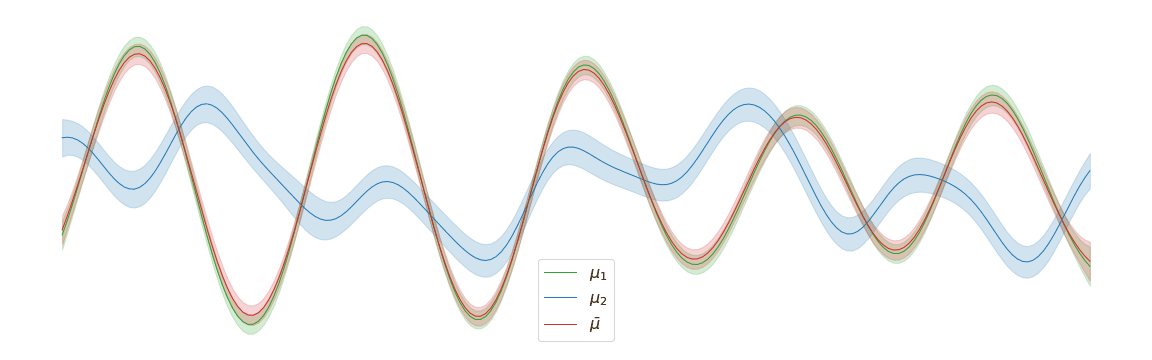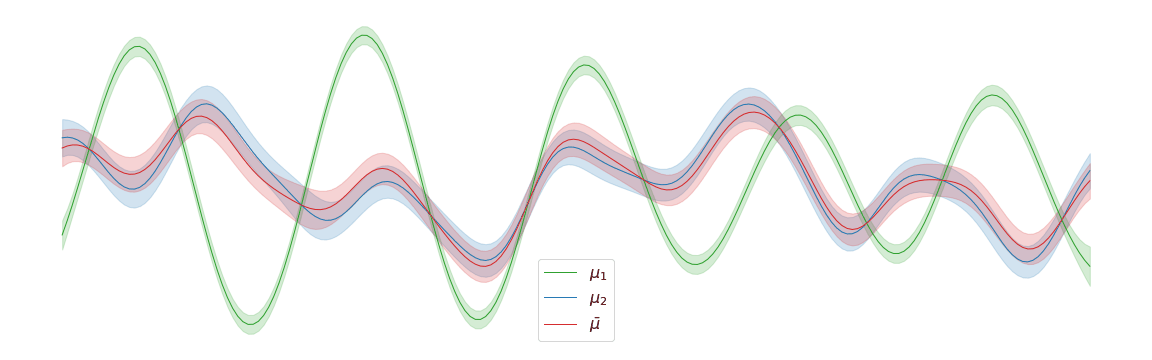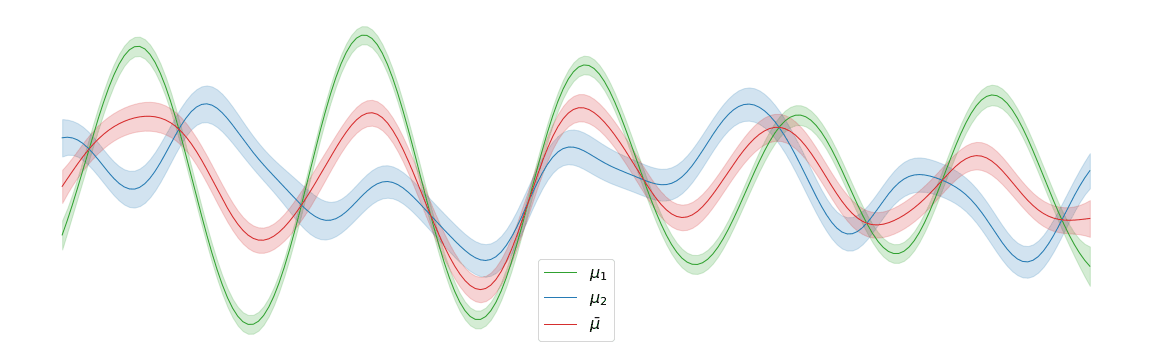
In the above example, we assigned uniform weights to each of the posteriors within the barycentre. In practice, we may have prior knowledge of which posterior is most likely to be the correct one. Regardless of the weights chosen, the barycentre remains a Gaussian process. We can interpolate between a pair of posterior distributions and to visualise the corresponding barycentre .
System configuration
Author: Thomas Pinder
Last updated: Mon Dec 15 2025
Python implementation: CPython
Python version : 3.11.14
IPython version : 9.5.0
gpjax : 0.13.4
matplotlib: 3.10.6
jaxtyping : 0.3.2
jax : 0.7.1
numpyro : 0.19.0
Watermark: 2.5.0